biglasso extends lasso and elastic-net linear and logistic regression models for ultrahigh-dimensional, multi-gigabyte data sets that cannot be loaded into memory. It utilizes memory-mapped files to store the massive data on the disk and only read those into memory whenever necessary during model fitting. Moreover, some advanced feature screening rules are proposed and implemented to accelerate the model fitting. As a result, this package is much more memory- and computation-efficient and highly scalable as compared to existing lasso-fitting packages such as glmnet and ncvreg. Bechmarking experiments using both simulated and real data sets show that biglasso is not only 1.5x to 4x times faster than existing packages, but also at least 2x more memory-efficient. More importantly, to the best of our knowledge, biglasso is the first R package that enables users to fit lasso models with data sets that are larger than available RAM, thus allowing for powerful big data analysis on an ordinary laptop.
Installation
To install the latest stable release version from CRAN:
install.packages("biglasso")To install the latest development version from GitHub:
remotes::install_github("pbreheny/biglasso")News
- See NEWS.md for latest news.
- The technical paper of this package was selected as a Winner of 2017 ASA Student Paper Competiton from Section on Statistical Computing.
- This package finished in the top 3 for 2017 ASA Chambers Statistical Software Award.
Documentation
- Here are the R Reference manual and Package Website
- Here are the technical papers of the package: i) The software paper; and ii) the paper of hybrid safe-strong rules
Features
- It utilizes memory-mapped files to store the massive data on the disk, only loading data into memory when necessary during model fitting. Consequently, it’s able to seamlessly handle out-of-core computation.
- It is built upon pathwise coordinate descent algorithm with warm start, active set cycling, and feature screening strategies, which has been proven to be one of fastest lasso solvers.
- We develop new, adaptive feature screening rules that outperform state-of-the-art screening rules such as the sequential strong rule (SSR) and the sequential EDPP rule (SEDPP) with additional 1.5x to 4x speedup.
- The implementation is designed to be as memory-efficient as possible by eliminating extra copies of the data created by other R packages, making
biglassoat least 2x more memory-efficient thanglmnet. - The underlying computation is implemented in C++, and parallel computing with OpenMP is also supported.
Benchmarks:
Simulated data:
-
Packages to be compared:
biglasso (1.4-0),glmnet (4.0-2),ncvreg (3.12-0), andpicasso (1.3-1). - Platform: AMD Ryzen 5 5600X @ 4.2 GHz and 32 GB RAM.
-
Experiments: solving lasso-penalized linear regression over the entire path of 100
lambdavalues equally spaced on the log scale oflambda / lambda_maxfrom 0.1 to 1; varying number of observationsnand number of featuresp; 20 replications, the mean computing time (in seconds) are reported. -
Data generating model:
y = X * beta + 0.1 eps, whereXandepsare i.i.d. sampled fromN(0, 1).
(1) biglasso is more computation-efficient:
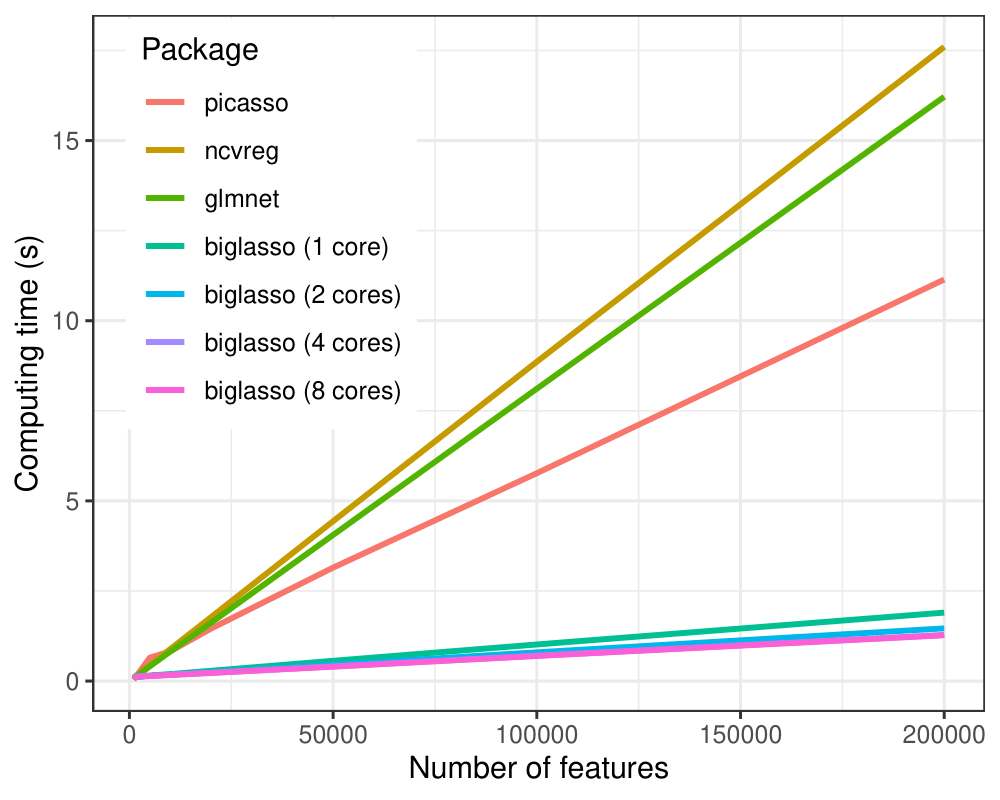
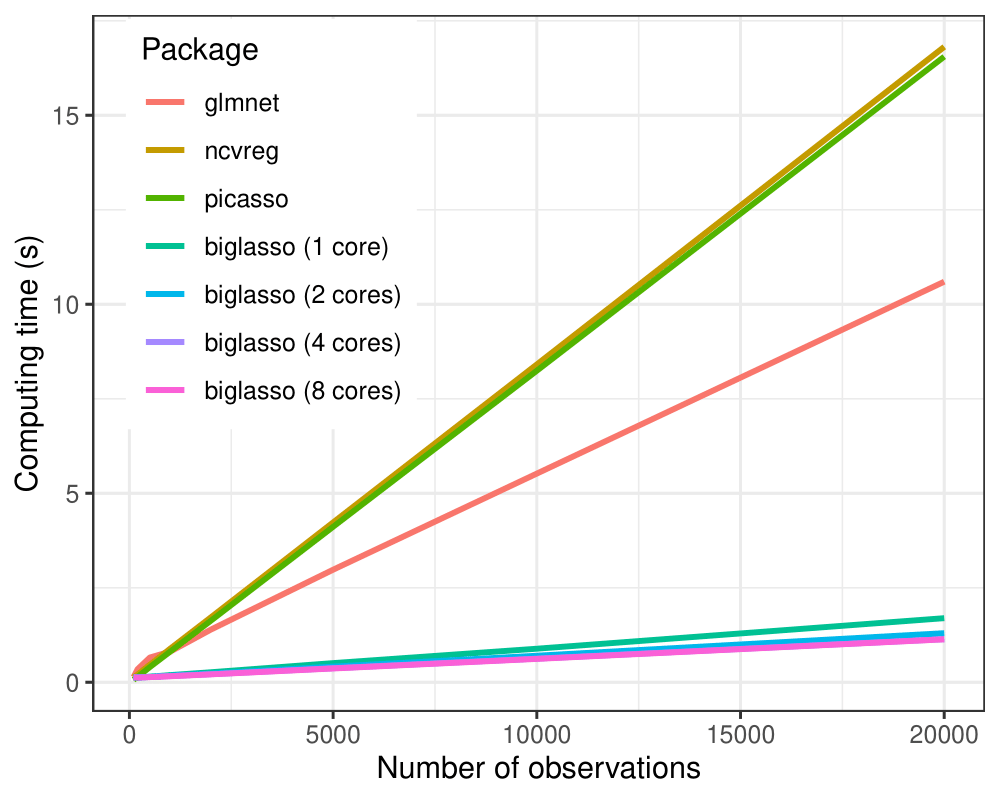
In all the settings, biglasso (1 core) is uniformly faster than picasso, glmnet and ncvreg. When the data gets bigger, biglasso achieves 6-9x speed-up compared to other packages. Moreover, the computing time of biglasso can be further reduced by half via parallel-computation of multiple cores.
(2) biglasso is more memory-efficient:
To prove that biglasso is much more memory-efficient, we simulate a 1000 X 100000 large feature matrix. The raw data is 0.75 GB. We used Syrupy to measure the memory used in RAM (i.e. the resident set size, RSS) every 1 second during lasso model fitting by each of the packages.
The maximum RSS (in GB) used by a single fit and 10-fold cross validation is reported in the Table below. In the single fit case, biglasso consumes 0.60 GB memory in RAM, 23% of that used by glmnet and 24% of that used by ncvreg. Note that the memory consumed by glmnet and ncvreg are respectively 3.4x and 3.3x larger than the size of the raw data. biglasso also requires less additional memory to perform cross-validation, compared other packages. For serial 10-fold cross-validation, biglasso requires just 31% of the memory used by glmnet and 11% of that used by ncvreg, making it 3.2x and 9.4x more memory-efficient compared to these two, respectively.
| Package | picasso | ncvreg | glmnet | biglasso |
|---|---|---|---|---|
| Single fit | 0.74 | 2.47 | 2.57 | 0.60 |
| 10-fold CV | - | 4.62 | 3.11 | 0.96 |
Note: ..* the memory savings offered by biglasso would be even more significant if cross-validation were conducted in parallel. However, measuring memory usage across parallel processes is not straightforward and not implemented in Syrupy; ..* cross-validation is not implemented in picasso at this point.
Real data:
The performance of the packages are also tested using diverse real data sets: * Breast cancer gene expression data (GENE); * MNIST handwritten image data (MNIST); * Cardiac fibrosis genome-wide association study data (GWAS); * Subset of New York Times bag-of-words data (NYT).
The following table summarizes the mean (SE) computing time (in seconds) of solving the lasso along the entire path of 100 lambda values equally spaced on the log scale of lambda / lambda_max from 0.1 to 1 over 20 replications.
| Package | GENE | MNIST | GWAS | NYT |
|---|---|---|---|---|
n=536 |
n=784 |
n=313 |
n=5,000 |
|
p=17,322 |
p=60,000 |
p=660,495 |
p=55,000 |
|
| picasso | 0.67 (0.02) | 2.94 (0.01) | 14.96 (0.01) | 15.91 (0.16) |
| ncvreg | 0.87 (0.01) | 4.22 (0.00) | 19.78 (0.01) | 25.59 (0.12) |
| glmnet | 0.74 (0.01) | 3.82 (0.01) | 16.19 (0.01) | 24.94 (0.16) |
| biglasso | 0.31 (0.01) | 0.61 (0.02) | 4.82 (0.01) | 5.91 (0.78) |
Big data: Out-of-core computation
To demonstrate the out-of-core computing capability of biglasso, a 96 GB real data set from a large-scale genome-wide association study is analyzed. The dimensionality of the design matrix is: n = 973, p = 11,830,470. Note that the size of data is 3x larger than the installed 32 GB of RAM.
Since other three packages cannot handle this data-larger-than-RAM case, we compare the performance of screening rules SSR and Adaptive based on our package biglasso. In addition, two cases in terms of lambda_min are considered: (1) lam_min = 0.1 lam_max; and (2) lam_min = 0.5 lam_max, as in practice there is typically less interest in lower values of lambdafor very high-dimensional data such as this case. Again the entire solution path with 100 lambda values is obtained. The table below summarizes the overall computing time (in minutes) by screening rule SSR (which is what other three packages are using) and our new rule Adaptive. (No replication is conducted.)
| Cases | SSR | Adaptive |
|---|---|---|
lam_min / lam_max = 0.1, 1 core |
189.67 | 66.05 |
lam_min / lam_max = 0.1, 4 cores |
86.31 | 46.91 |
lam_min / lam_max = 0.5, 1 core |
177.84 | 24.84 |
lam_min / lam_max = 0.5, 4 cores |
85.67 | 15.14 |
Reference:
- Zeng Y and Breheny P (2021). The biglasso Package: A Memory- and Computation-Efficient Solver for Lasso Model Fitting with Big Data in R. R Journal, 12: 6-19. URL https://doi.org/10.32614/RJ-2021-001
- Zeng Y, Yang T, and Breheny P (2021). Hybrid safe-strong rules for efficient optimization in lasso-type problems. Computational Statistics and Data Analysis, 153: 107063. URL https://doi.org/10.1016/j.csda.2020.107063
- Wang C and Breheny P (2022). Adaptive hybrid screening for efficient lasso optimization. Journal of Statistical Computation and Simulation, 92: 2233–2256. URL https://doi.org/10.1080/00949655.2021.2025376
- Tibshirani, R., Bien, J., Friedman, J., Hastie, T., Simon, N., Taylor, J., and Tibshirani, R. J. (2012). Strong rules for discarding predictors in lasso-type problems. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 74 (2), 245-266.
- Wang, J., Zhou, J., Wonka, P., and Ye, J. (2013). Lasso screening rules via dual polytope projection. In Advances in Neural Information Processing Systems, pp. 1070-1078.
- Xiang, Z. J., and Ramadge, P. J. (2012, March). Fast lasso screening tests based on correlations. In Acoustics, Speech and Signal Processing (ICASSP), 2012 IEEE International Conference on (pp. 2137-2140). IEEE.
- Wang, J., Zhou, J., Liu, J., Wonka, P., and Ye, J. (2014). A safe screening rule for sparse logistic regression. In Advances in Neural Information Processing Systems, pp. 1053-1061.
Report bugs:
- open an issue or send an email to Patrick Breheny at patrick-breheny@uiowa.edu